Signalling in hESCs and hiPSCs cells derived from human fibroblasts
In hESCs (human embryonic stem cells) and hiPSCs (human induced pluripotent stem cells) the TGFß signaling pathway is crucial for maintaining self-renewal and pluripotency (Babaie et al. 2007, Greber et al, 2007; Greber et al., 2008). In fact the TGFß/ACTIVIN/NODAL axis of the pathway needs to be activated via phosphorylation of SMAD2,3 and the BMP axis repressed via the non-phosphorylation of SMAD1,5,8 in order to sustain self-renewal and prevent differentiation (see figure 1 taken from Greber et al. 2007). 
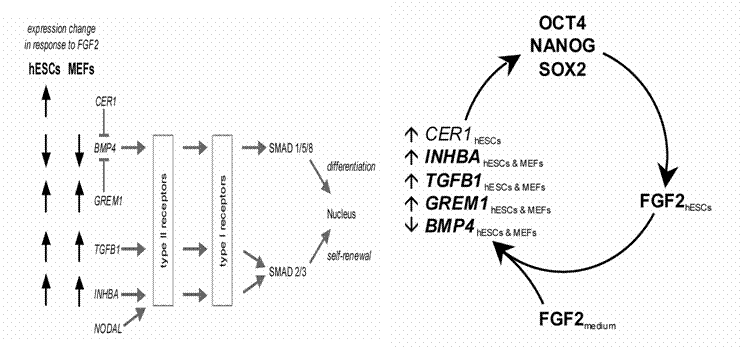
Figure 1: The growth factor, FGF2 regulates the expression of key members of the TGFß pathway with ACTIVIN-A, TGFß1, GREM1, and BMP4 being the most likely candidates encoding the above activities (Greber et al. 2007). The genes GREM1, CER1 and GDF3 are expressed in undifferentiated hESCs and hiPSCs, and they encode proteins known to act as antagonist of BMP4 activity. Stimulation of hESCs or iPSCs with BMP2 or BMP4 induces differentiation to the trophoblast lineage.
The maintenance of pluripotency and self-renewal of human ES and iPS cells are intrinsically complex processes driven by the co-ordinated dynamic expression of a plethora of genes, their encoded proteins and associated signalling pathways in response to external signaling cues such as FGF2. Our systems biology approach combines high throughput approaches (OCT4 ChIP-chip, ChIP-seq, RNAi, protein interaction networks, metabolomics and cytokine stimulations of hESCs) and advanced computational techniques to dissect the molecular mechanisms of stem cell fate and cellular reprogramming (Fig.2).
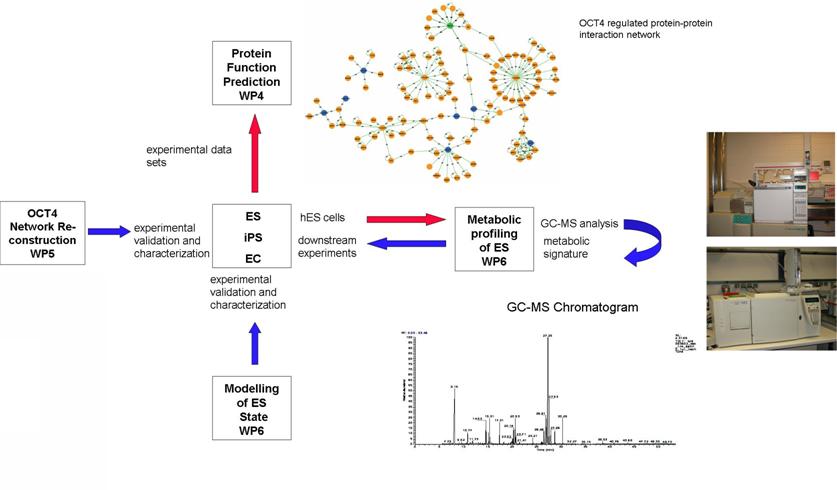
Figure 2: Network Reconstruction, analysis and network-based modeling of the human ES and iPS self-renewal Gene Regulatotry Network are carried out within the framework of the EU project “Experimental Network for Functional Integration” (ENFIN/FP6) Enabling Systems Biology.
Additionally, we have created the user-friendly Embryonic Stem Cell Database (ESCD) integrating available genomics related datasets in both human and mouse ES and EC cells (Fig. 3). In the current era of systems biology driven research, we envisage that our integrated embryonic stem cell database will prove beneficial to the booming field of ES, iPS and cancer research.
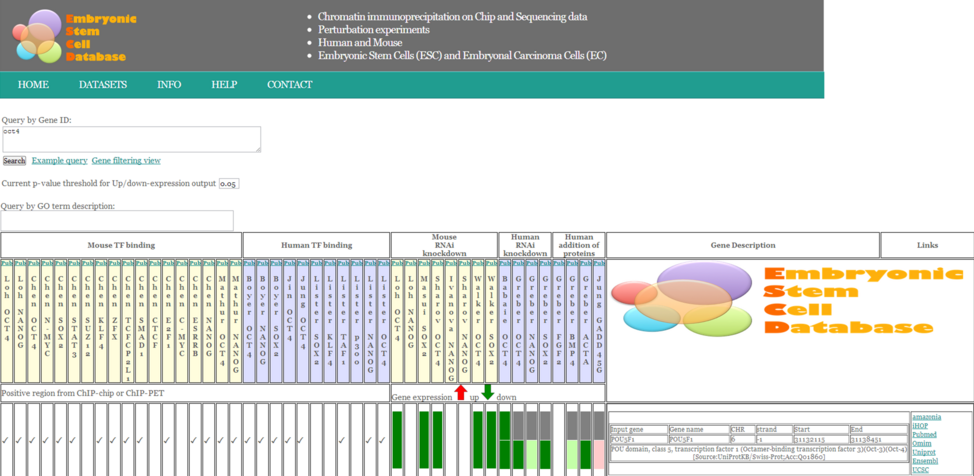
Figure 3: The Embryonic Stem Cell Database (Jung and Peterson et al.,2010)
In order to study gene regulatory networks that might be important for pluripotency and self-renewal of human ES and iPS cells we are using STRING network (v.9.0). An OCT4 gene regulatory network generated by STRING is illustrated in Fig. 4.
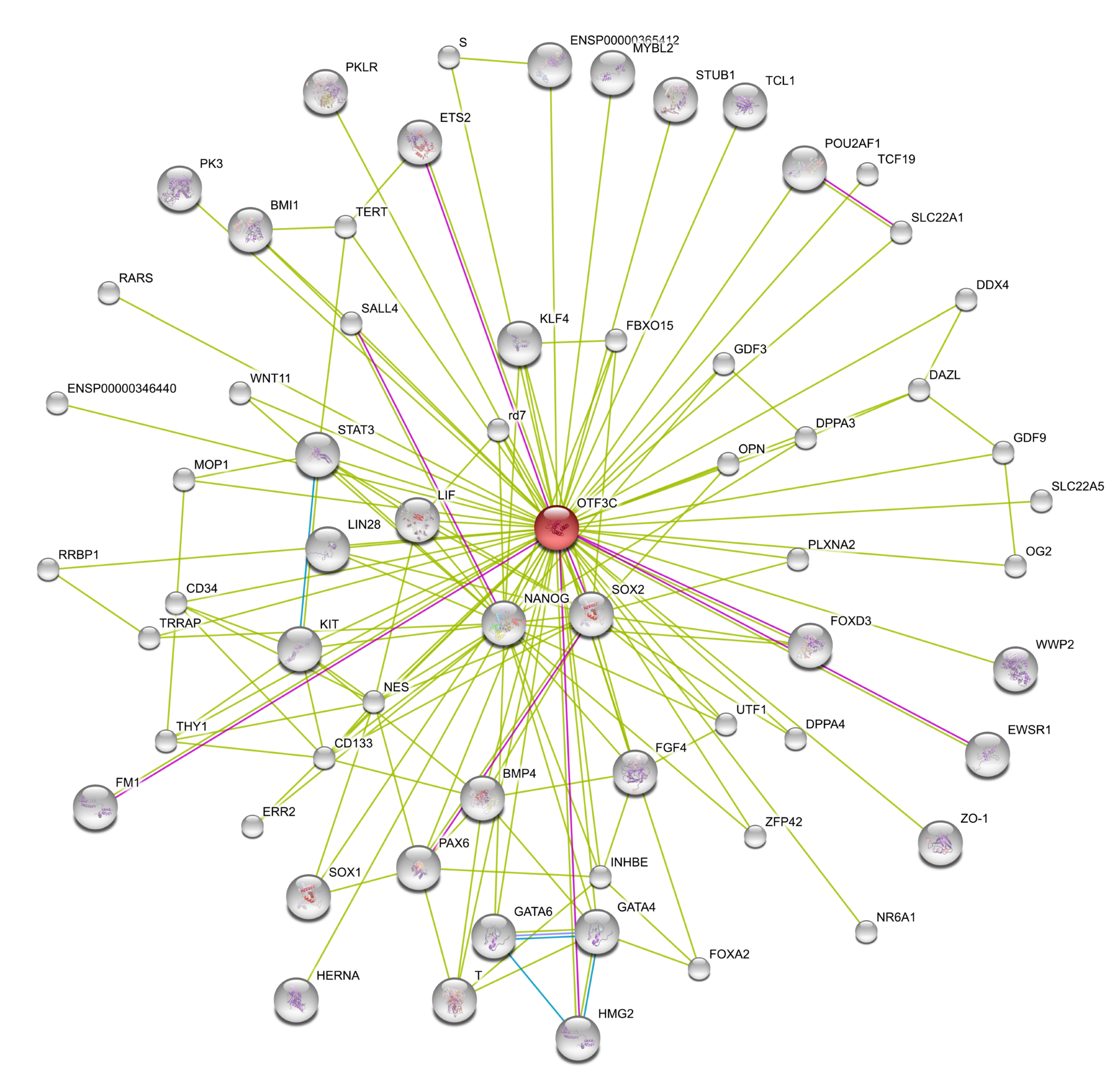
Figure 4: STRING network (v.8.0) based on only OCT4 /OTF3C (red) using high confidence, and network depth 2. Green line indicates links based on textmining, violet based on experiments.
References:
Control of early fate decisions in human ES cells by distinct states of TGFbeta pathway activity.
Greber B, Lehrach H, Adjaye J.
Stem Cells Dev. 2008 Dec;17(6):1065-77. PMID: 18393632 PubMed
A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells.
Jung M, Peterson H, Chavez L, Kahlem P, Lehrach H, Vilo J, Adjaye J.
PLoS One. 2010 May 21;5(5):e10709. PMID: 20505756 PubMed
Silencing of core transcription factors in human EC cells highlights the importance of autocrine FGF signaling for self-renewal.
Greber B, Lehrach H, Adjaye J.
BMC Dev Biol. 2007 May 16;7:46. PMID: 17506876 PubMed
Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells.
Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, Lehrach H, Burdon T, Adjaye J.
Stem Cells. 2007 Feb;25(2):500-10. Epub 2006 Oct 26. PMID: 17068183 PubMed




