Research
CORE PLATFORM
A functional kidney is essential for healthy living due to its major role in toxin and drug filtration. Globally, each year millions of patients require rapid kidney transplantation or dialysis to restore renal function. Furthermore, the shortage of compatible organs, donor-associated diseases, ageing-associated factors and high cost of transplantation/dialysis are major hurdles. Kidney associated dysfunctions are now a prioritized health concern and research area. A potential alternative source of renal cells are those derived from embryonic and induced pluripotent stem cells (ESCs and iPSCs). However, clinical applications of pluripotent stem cell technologies are constrained by the risk of tumor formation, immunological rejection, legal as well as ethical concerns.
In light of this, it is therefore important to find other sources of kidney stem cells which are not tumorigenic, bear a broad differentiation potential and have a high renal regenerative potential. It is estimated that approximately 2000 to 7000 cells are flushed out of the renal system in our urine, which contain podocytes, different epithelial cells, blood cells and some renal progenitor cells. Urine stem cells (USCs) originate from the metanephric mesenchyme and those from the glomeruli are capable of giving rise to podocytes, proximal tubular cells and distal cells. USCs express renal cell markers such as PAX2, PAX8, podocyte-related proteins- Synaptopodin, Nephrin, Podxl and Podocin
USCs exhibit stem cell properties such as high proliferation capacity as they express telomerase activity and have multi-differentiation potential. Like bonemarrow derived mesenchymal stem cells, USCs express similar surface markers -CD24, CD29, CD44, CD73, CD90, CD105, CD117, CD133, CD146, SSEA-4 and STRO-1, but not the hematopoietic stem cell markers CD14, CD31, CD34 and CD45. Due to their multi-differentiation potential, autologous urine derived stem cells can be safely transplanted for treating kidney-related diseases.
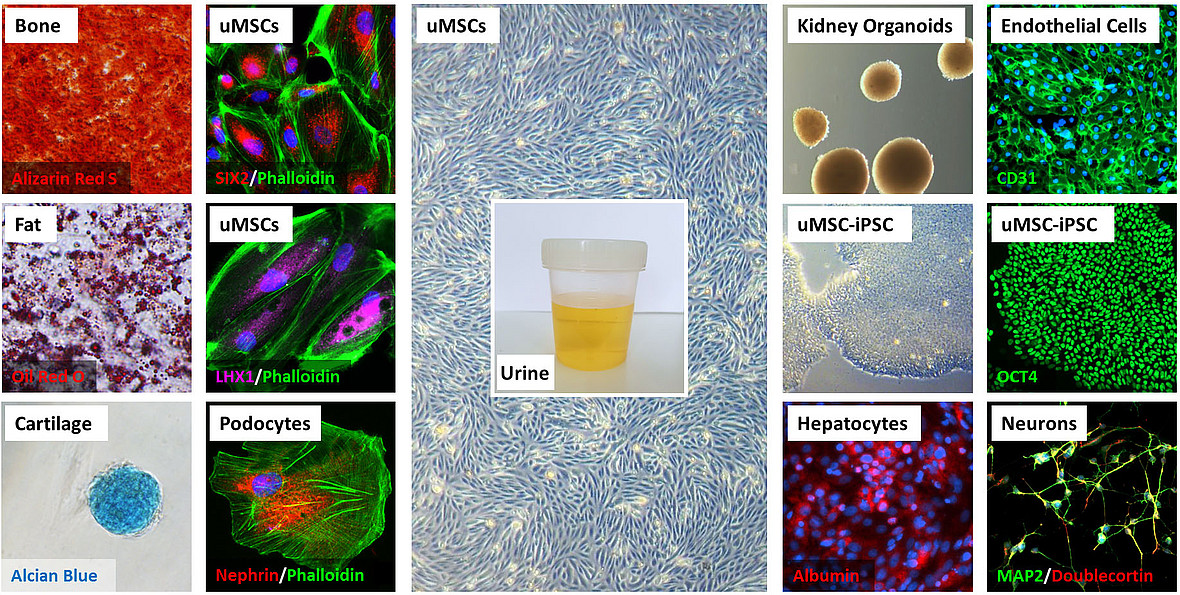
We use our urine platform to enable in vitro studies on nephrogenesis, kidney associated diseases and the derivation of iPSCs to enable further studies on liver and brain associated diseases.
Propagation and characterisation of urine-derived renal progenitors- UdRPCs.

(A) Representative pictures of the “rice grain”-like appearance of the cells from the initial attachment to an elongated MSC-like morphology.
(B) Growth curve analysis of selected urine-derived renal progenitors carried out using the Resazurin metabolic assay. Data are presented as means ± SEMs.
(C) Immune-phenotyping for SSEA4, TRA-1-81 and TRA-1-60.
(D) immunofluorescence-based detection of the expression of pluripotency-associated stem cell- proteins SSEA4 (red), C-KIT (green), CD133 (red) and the mesenchymal-associated protein Vimentin (green); cell nuclei were stained using Hoechst/DAPI (scale bars: 100 μm and 50 μm). (E) Bisulfite sequencing of CpG-island methylation patterns within the 5′- regulatory region of the OCT4 gene in UM51. Filled circles represent methylated CpG dinucleotides. White circles represent unmethylated CpGs. Arrows indicate the transcription start-site.
Expression of kidney-associated proteins in urine-derived renal progenitors and Albumin transport.

(A) Urine-derived renal progenitors express the renal stem cell markers- SIX2, CITED1, WT1, and CK19. Renal markers (red) and cell nuclei were stained using DAPI/Hoechst (blue). (B) Flow cytometry analysis for the key renal stem cell transcription factor SIX2 and (C) Renal stem cell surface markers CD24, CD133, and CD106 of UM27, UF31, UF45 and UM51. (D) Detailed CpG methylation profiles of the SIX2 5′-regulatory region are documented as revealed by bisulfite sequencing. Filled circles represent methylated CpG dinucleotides and white circles unmethylated CpGs. Arrows indicate the transcription start site. 1.9% of CpG dinucleotides were found to be methylated. (E) Urine-derived renal progenitors (n = 4) like the human kidney biopsy-derived hREPCs also transport Albumin. Albumin was coupled to Alexa Fluor 488 (green) and cell nuclei stained with DAPI (blue). Scale bars indicate 50 μm
Integration-free iPSCs can be efficiently derived from UdRPCs using episomal-based plasmids without pathway perturbation

Multi-Omics
Omics-based datasets (transcriptome, proteome, methylome and secretome) bioinformatics, mathematical modeling, pathway reconstruction and data management are central to our research.