Experimental Oncology and Immunology – Projects
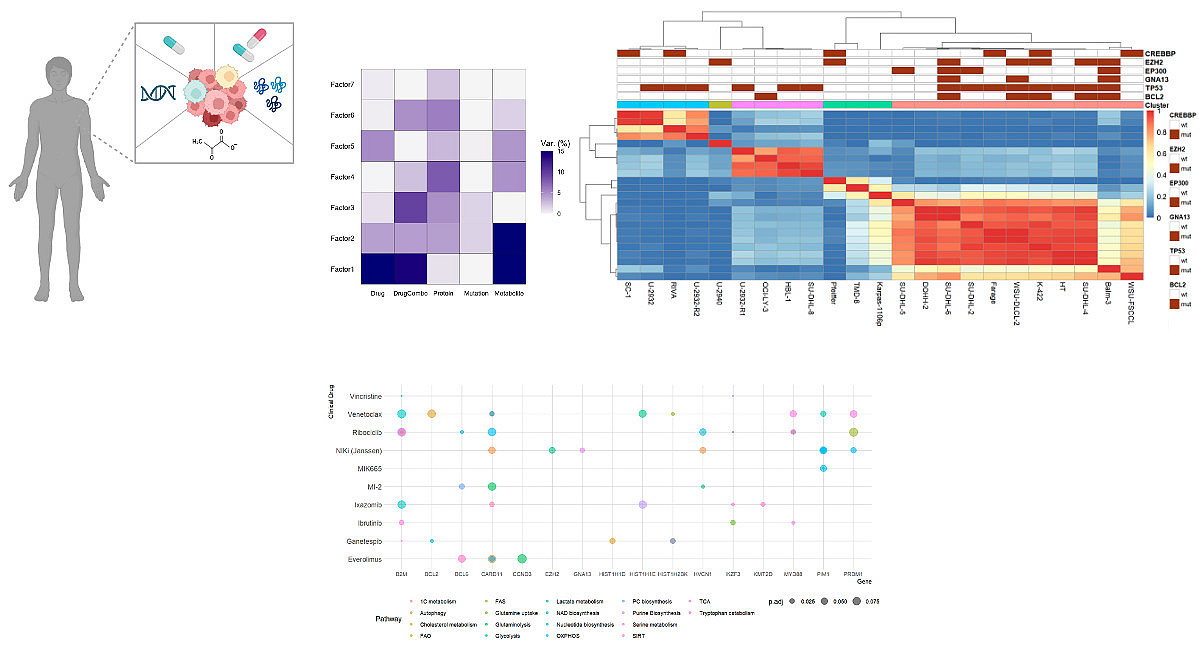
Metabolic Vulnerabilities in Diffuse Large B Cell Lymphoma (DLBCL) - This project focuses on unraveling intrinsic and acquired resistance mechanisms linked to metabolism in Diffuse Large B Cell Lymphoma. Probing resistance mechanisms by a systematic comparison of genomic, proteomic, and metabolomic information with ex-vivo drug screening responses allows us to identify metabolic alterations driving treatment resistance and design therapeutic approaches to overcome these resistance mechanisms.
Pathogenesis of Chronic Lymphocytic Leukemia (CLL) - This project aims to identify a collection of protein markers capable to stratify CLL patient outcome through daily routine methods (FACS/qRT-PCR). Starting from proteomics biomarkers, previously identified to be associated with inferior outcome CLL, this project emphasizes the development of a combined FACS panel/qRT-PCR splice variant screening to identify high risk CLL patients early.
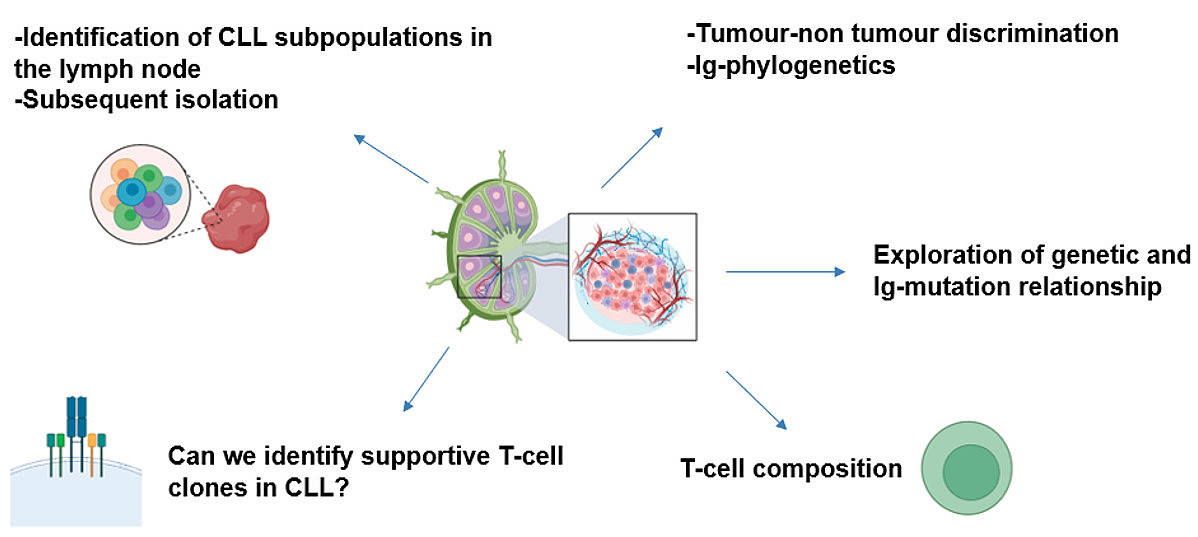
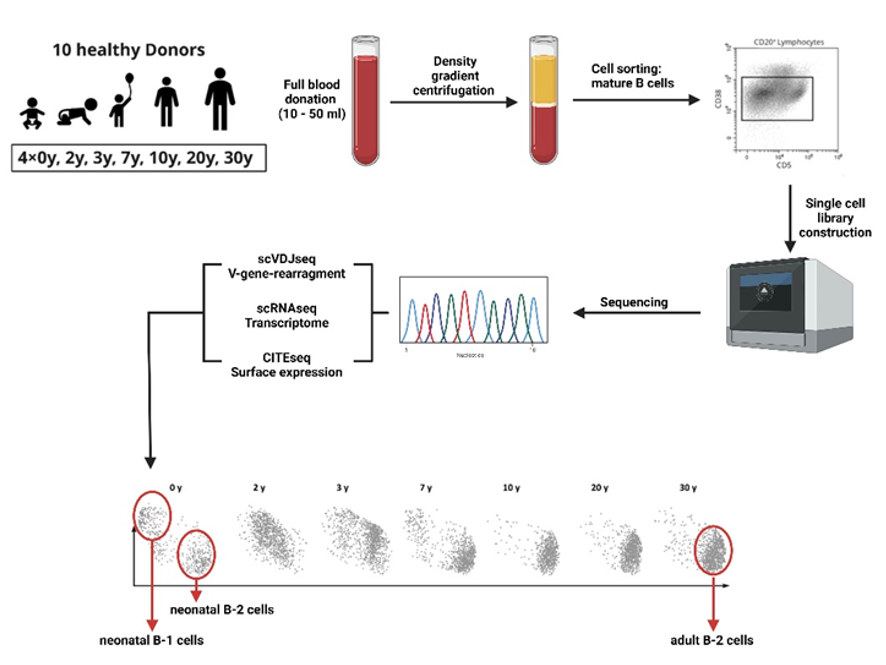
Lymphoma Originating Cells in CLL - This project has the goal to identify putative CLL originating cells in the lymph nodes through single cell and bulk sequencing approaches (WGS-RNA/VDJ/CITEseq). This study is supposed to unravel different CLL subpopulations and their hierarchical and pathogenetic relationship. Moreover, we aim to investigate T cell heterogeneity in the CLL lymph node microenvironment.
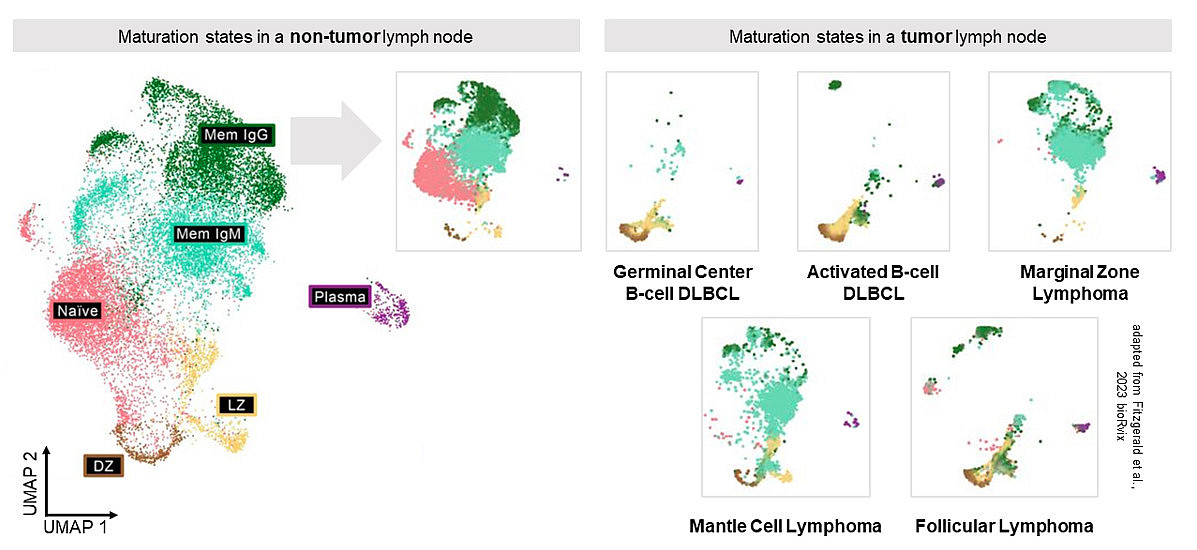
Lymphoma Heterogeneity - Our research is dedicated to unravel the complexities of tumor heterogeneity within B-cell lymphomas. We focus on the development and characterization of subpopulations present in primary patient samples, utilizing techniques such as RNA-seq, CITE-seq, and VDJ-seq.
Immune Receptor Repertoires in Endemic Burkitt‘s Lymphoma (eBL) - Our objective is to explore the individual contributions of Malaria and Epstein-Barr Virus in the pathogenesis of endemic Burkitt Lymphoma (eBL). Through profiling the T- and B-cell receptor repertoires in mouse models, we seek to reveal how each infectious agent distinctly – and in coinfection - perturbs the dynamics of the germinal center and sustains lymphomagenesis.
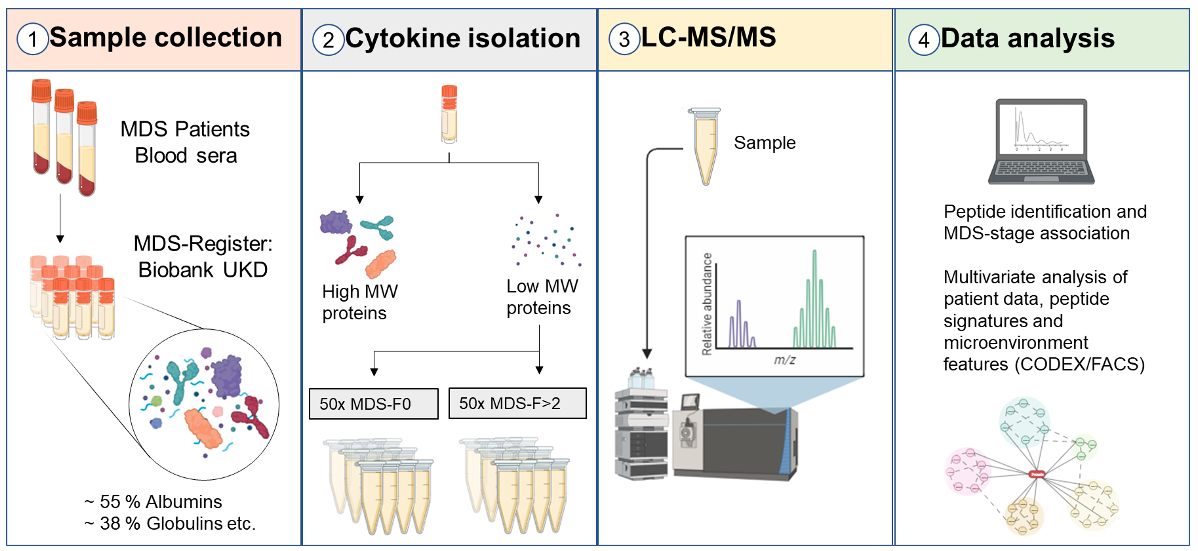
Cytokine Networks in Myelodysplastic Syndrome (MDS) - We aim to investigate the cytokine network in myelodysplastic syndrome (MDS), a clonal disorder of hematopoietic stem cells, to identify patterns useful for diagnostic and prognostic questions. Prediction of the disease progression is crucial to stratify treatment. A particular focus of this project is to understand the impact of fibrosis in MDS.
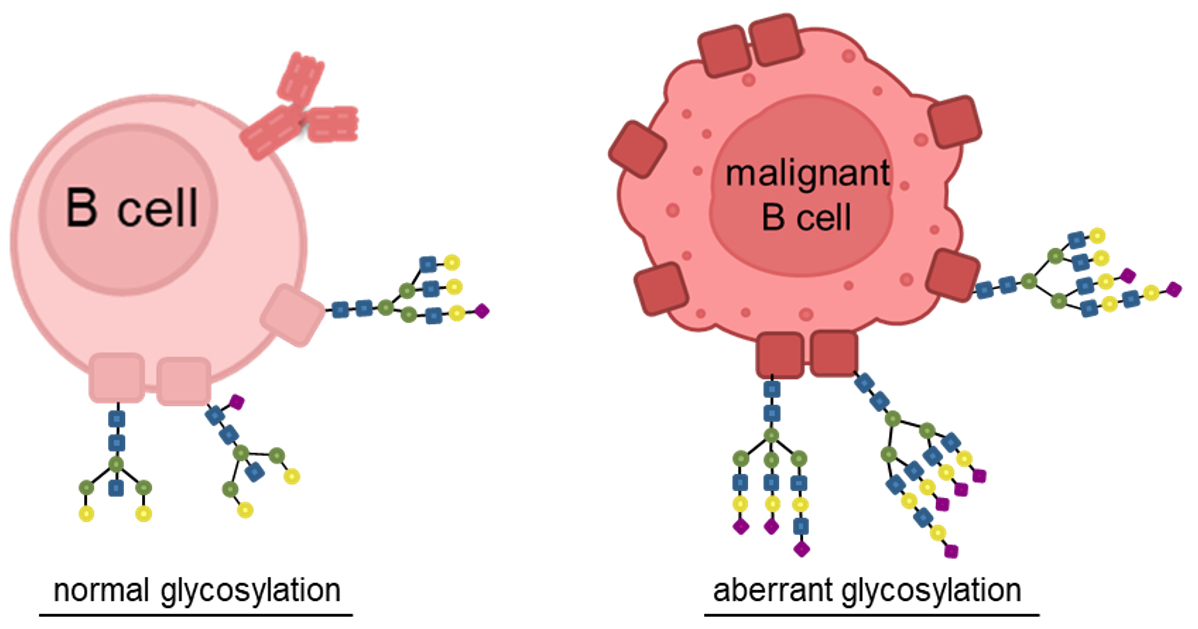
Glycoproteomics and Tumor Antigen Profiling - This project aims at investigating the glycosylation patterning and homeostasis of surface proteins in lymphoma entities. By understanding alterations in glycan patterns, we aim to develop strategies to overcome immunotherapeutic resistances and enhance treatment efficacy. Our goal is to use the insights of glycoproteomic to advance personalized immune therapies and improve outcomes for patients with lymphoma.
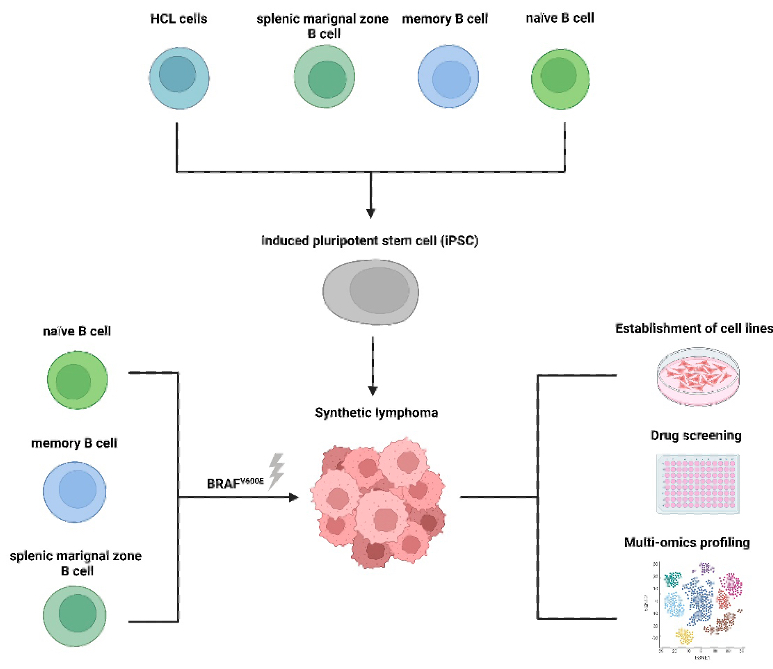
Synthetic Lymphomas - Hairy Cell Leukemia (HCL) Pathogenesis - Our aim is to explore interactions of Hairy cell leukemia (HCL) and the bone marrow microenvironment, discover metabolomic vulnerabilities of HCL and consequences for drug response to disrupt pro-survival pathways as therapeutic opportunities. To achieve this, we established a systems medicine program that integrates functional assays, multi-omics profiling and bioinformatic analysis.
We further aim to reconstruct HCL pathogenesis by generating a synthetic lymphoma cell line.