Personalized Drug Treatment - Projects
We aim to leverage cutting-edge technology in conjunction with patient-derived primary cancer cells to determine individualized drug responses. Additionally, we will integrate analyses of genetic, epigenetic, transcriptomic, and proteomic factors to identify potential treatment options. By employing this comprehensive approach, we aim to provide a more personalized and effective treatment plan for cancer patients.
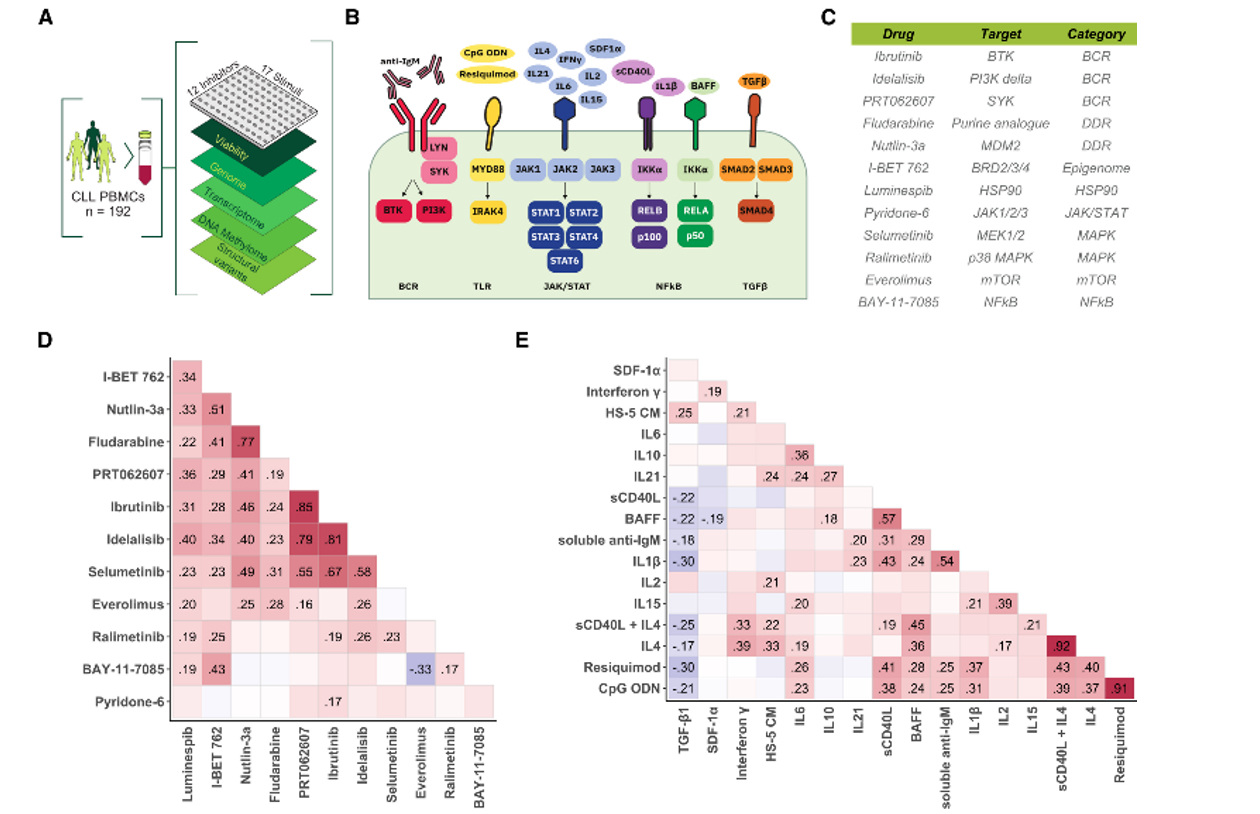
Drug response profiling (DRP) - Treatment response in cancer is influenced by multiple biological factors including genetic alterations of tumor cells and the tumor microenvironment. To investigate modulators of therapy response in patients, we can model drug response ex vivo. For this, we isolate primary tumor cells from peripheral blood or diagnostic biopsies and treat them with a large library of clinically available and experimental drugs.
Additionally to important biological findings, ex vivo DRP can be used to predict the response of individual patients beyond known markers of therapy resistance. Based on this readout, we were able to treat patients who lacked any further standard treatment options.
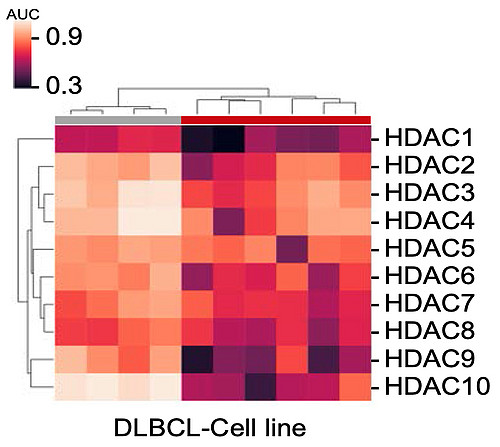
Histone Deacetylase (HDAC) Inhibitor Drug Screening in Diffuse Large B cell Lymphoma (DLBCL) -
Working on a close collaboration project with the Institute for Pharmaceutical and Medicinal Chemistry. In this project, the specific targeted effect of the newly developed histone deacetylase is investigated.
Live Cell Imaging - This project involves a collaborative effort with the Chair in Algorithmic Bioinformatics to characterize the dynamic response to drugs using a live-cell imaging-based detection and computational modelling approach.