What we do
We pursue four specific objectives:
1.) The development of deep learning methods for diagnosis and risk prediction of liver diseases based on histological image data, including multicentre blinded validation:
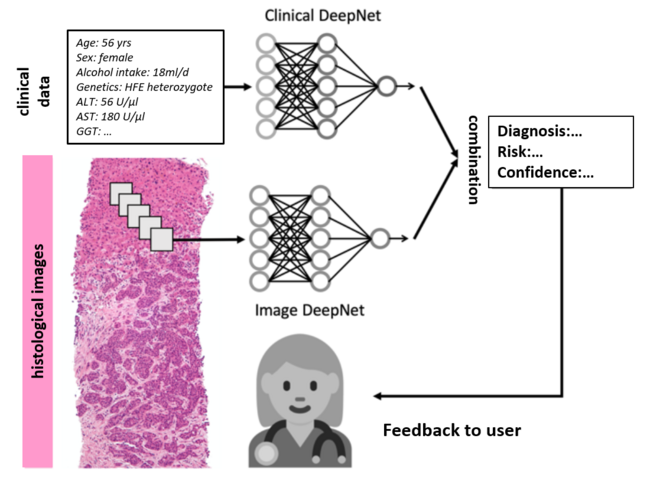
We develop deep learning systems that automatically pre-process histological image data from routine liver biopsies and perform quality control and colour normalisation and generate small, overlapping image areas ("tiles") with which a deep neural network is trained. This Image DeepNet is trained to diagnose the patient's disease and predict the clinical course. This diagnostic tool is validated in a blinded fashion using large, multicentre patient cohorts.
The deep learning tools developped by Jakob Kather and his team are published on github: Kather Lab at EKFZ / TU Dresden · GitHub
2.) Systematic comparison of image-based risk prediction with clinical data and classical risk scores as well as development of multimodal deep learning systems:
Deep learning networks are trained on routine clinical data to diagnose and predict clinical endpoints (Clinical DeepNet). The performance of this methodology is compared with pure image-based data analysis. Subsequently, the information from the Clinical DeepNet is combined with the Image DeepNet and summarised using classical machine learning models.
3.) Explaining statistically predictive features and finding novel clusters and previously unknown biological subgroups of liver diseases based on unsupervised methods.
In a "reverse engineering" step, the features in the data set are identified which are particularly associated with the target variable. In image data, this could be, for example, specific tissue components such as connective tissue septa or immune cell infiltrates in liver tissue. This process may reveal new biological properties associated with specific diseases or clinical courses and thus fundamentally expand our understanding of liver diseases.
4.) The development of a user-friendly and intuitive web-based platform for the broad dissemination of the developed methods and their systematic evaluation.
The long-term goal of our consortium is to introduce novel diagnostic and prognostic methods into mainstream medical practice. We developped a prototype web application in which scanned histological images in anonymised form can be uploaded, which are then automatically analysed. Based on this, a heatmap of the model prediction is generated:
Follow this link to find four different whole slide images from the TCGA database (The Cancer Genome Atlas Program), each with predictions from a model differentiating hepatocellular from cholangiocarcinoma.